modos.genomics.htsget#
htsget client implementation
The htsget protocol [1] allows to stream slices of genomic data from a remote server. The client is implemented as a file-like interface that lazily streams chunks from the server.
In practice, the client sends a request for a file with a specific format and genomic region. The htsget server finds the byte ranges on the data server (e.g. S3) corresponding to the requests and responds with a “ticket”.
The ticket is a json document containing a list of blocks; each having headers and a URL pointing to_file the corresponding byte ranges on the data server.
The client then streams data from these URLs, effectively concatenating the blocks into a single stream.
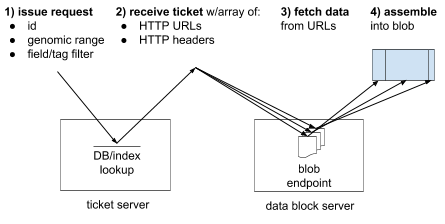
Illustration of the mechanism through which the htsget server allows streaming and random-access on genomic files. See [1] for more details.#
Notes
This implementation differs from the reference GA4GH implementation [2] in that it allows lazily consuming chunks from a file-like interface without saving to a file. A downside of this approach is that the client cannot seek.
Additionally, this implementation does not support asynchronous fetching of blocks, which means that blocks are fetched sequentially.
References
Classes#
Transparent iterator over blocks of an htsget stream. |
|
A file-like handle to a read-only, buffered htsget stream. |
|
Connection to an htsget resource. |
Functions#
|
Build an htsget URL from a host, path, and region. |
|
Given a URL to an htsget resource, extract the host, path, and region. |
Module Contents#
- modos.genomics.htsget.build_htsget_url(host, path, region)[source]#
Build an htsget URL from a host, path, and region.
Examples
>>> build_htsget_url( ... "http://localhost:8000", ... Path("file.bam"), ... Region("chr1", 0, 1000) ... ) 'http://localhost:8000/reads/file?format=BAM&referenceName=chr1&start=0&end=1000'
- Parameters:
host (pydantic.HttpUrl)
path (pathlib.Path)
region (modos.genomics.region.Region | None)
- Return type:
- modos.genomics.htsget.parse_htsget_url(url)[source]#
Given a URL to an htsget resource, extract the host, path, and region.
- Parameters:
url (pydantic.HttpUrl)
- Return type:
tuple[str, pathlib.Path, modos.genomics.region.Region | None]
- class modos.genomics.htsget._HtsgetBlockIter(blocks, chunk_size=65536, timeout=60)[source]#
Transparent iterator over blocks of an htsget stream.
This is used internally by HtsgetStream to lazily fetch and concatenate blocks.
Examples
>>> next(_HtsgetBlockIter([ ... {"url": "data:;base64,MTIzNDU2Nzg5"}, ... {"url": "data:;base64,MTIzNDU2Nzg5"}, ... ])) b'123456789'
- class modos.genomics.htsget.HtsgetStream(blocks)[source]#
Bases:
io.RawIOBaseA file-like handle to a read-only, buffered htsget stream.
Examples
>>> stream = HtsgetStream([ ... {"url": "data:;base64,MTIzNDU2Nzg5Cg=="}, ... {"url": "data:;base64,MTIzNDU2Nzg5Cg=="}, ... ]) >>> stream.read(4) b'1234'
- readable()[source]#
Return whether object was opened for reading.
If False, read() will raise OSError.
- Return type:
- readinto(b)[source]#
Read up to len(b) bytes into a writable buffer bytes and return the number of bytes read.
Notes
See https://docs.python.org/3/library/io.html#io.RawIOBase.readinto
- Parameters:
- Return type:
- class modos.genomics.htsget.HtsgetConnection[source]#
Connection to an htsget resource. It allows to open a stream to the resource and lazily fetch data from it.
- path: pathlib.Path[source]#
- region: modos.genomics.region.Region | None[source]#
- to_file(path)[source]#
Save all data from the stream to a file.
- Parameters:
path (pathlib.Path)
- classmethod from_url(url)[source]#
Open connection directly from an htsget URL.
- Parameters:
url (str)
- to_pysam(reference_filename=None)[source]#
Convert the stream to a pysam object.
- Parameters:
reference_filename (str | None)
- Return type:
collections.abc.Iterator[pysam.AlignedSegment | pysam.VariantRecord]
